Results
Hierarchical Normalized Cuts
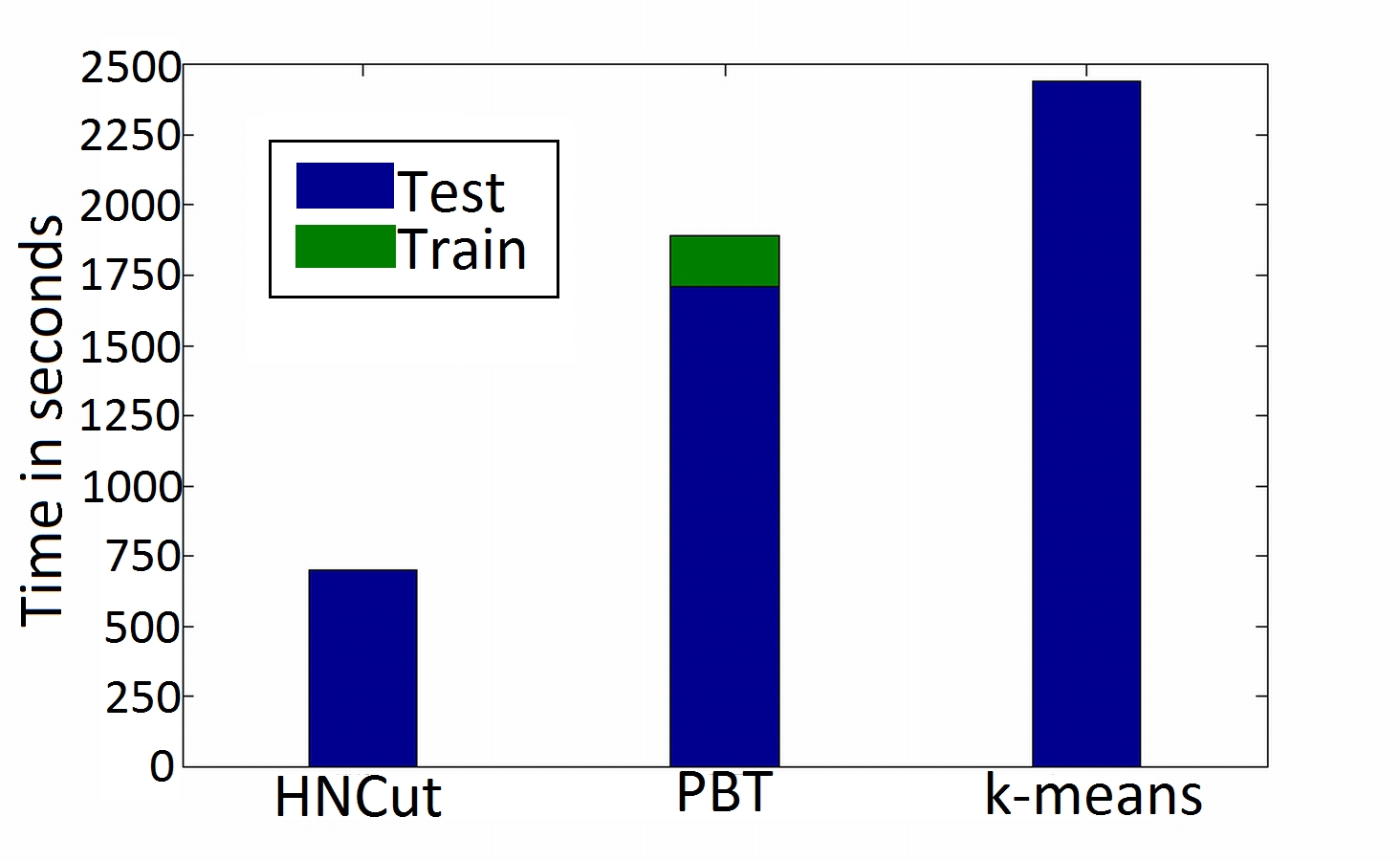
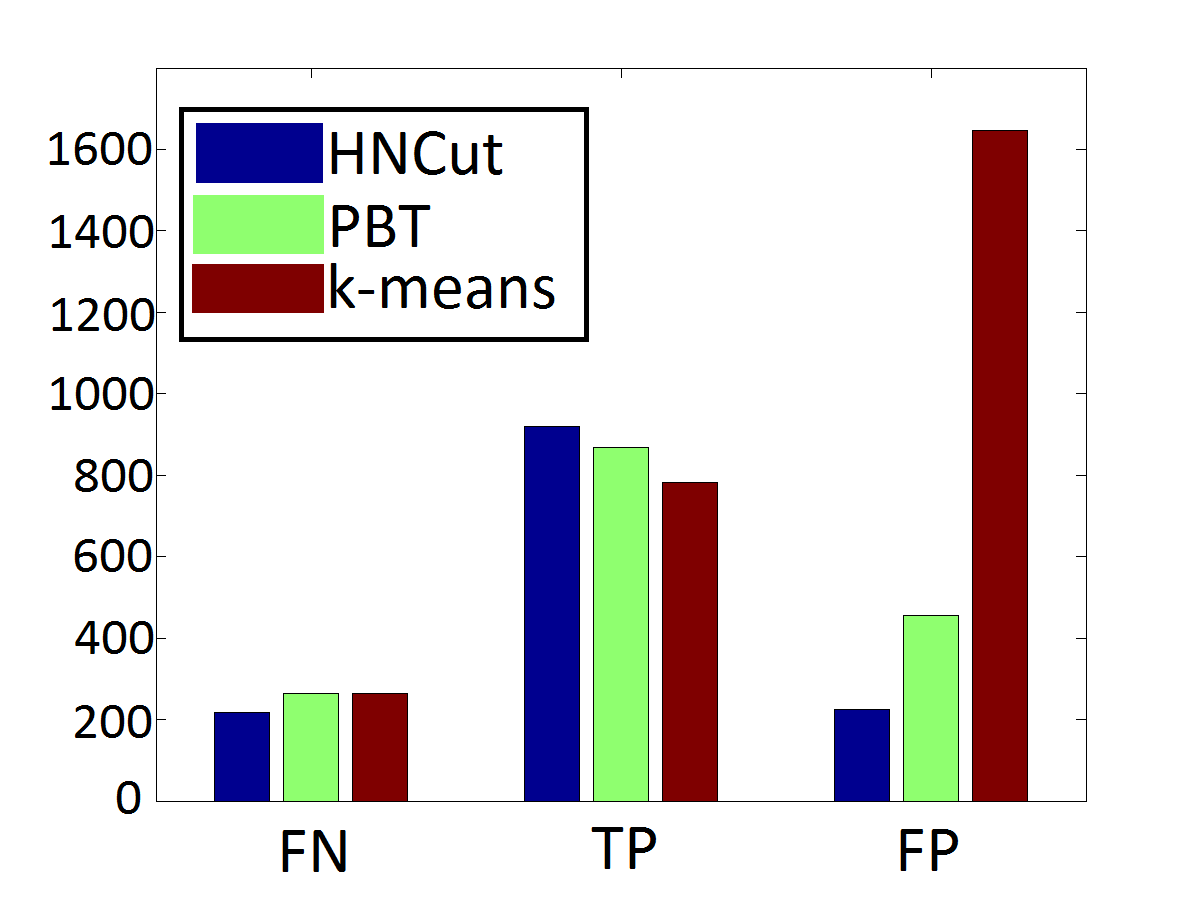
As an example of the value of HNCut, we present a comparison against a classical k-means approach and a powerful supervised classifier termed Probablistic Boosting Trees. As we can see, in both accuracy and computation time, HNCut is a superior algorithm at this segmentation task.
Local Morphometric Scale
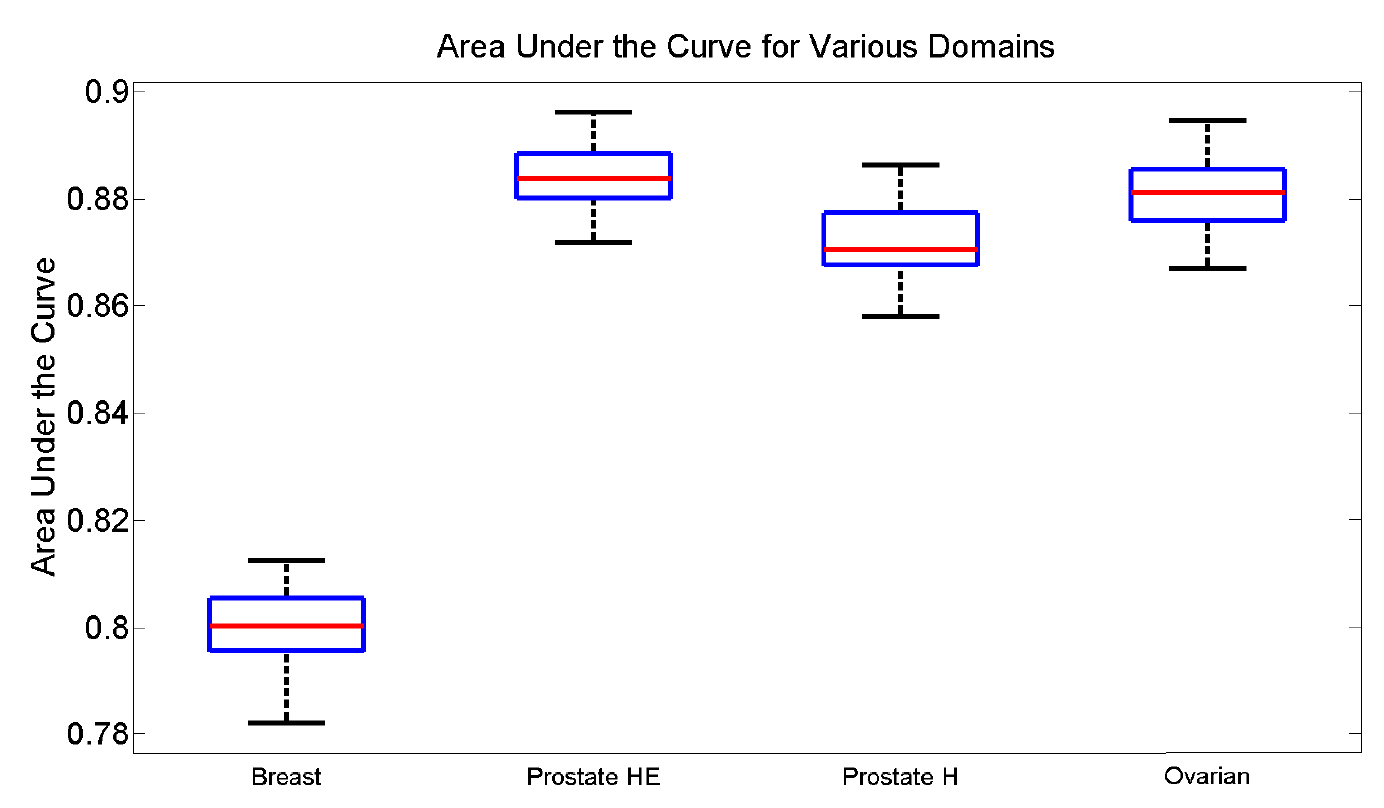
In the domain of classification, we aim to provide a signature at the pixel level which can be used to successfully differentiate tumor regions from stromal regions. The the chaotic nature of the regionions of interest (e.g. size and shape) prevents the selection of optimal operating parameters for standard industry algorithms, such as window size for texture features. We note, however, that the technique developed here is not specific to only tumor and stroma classification.
For four data domains, we randomly sampled pixels, broke them into training and test sets and used a naive Bayesian classifier to perform the classification. From the ROC curve, we computed the mean area under the curve (AUC) and associated range. What is especially interesting to note is that the parameters for each data domain were kept constant, which we feel indicates a strong non-domain specific feature set.