Summary
Hierarchical Normalized Cuts
The major contribution of this piece of the thesis is a fast, novel, hierarchical unsupervised segmentation method (HNCut), which we demonstrate with an application in Ovarian tissue micro arrays (TMA).Challenges
- Datasets are very large and thus require a highly efficient algorithm to make computation tractable. Additionally, a large number of these datasets already exist in tissue repositories waiting to be mined.
- Images are not consistent across the dataset due to lighting, staining, and human preparation variations. This anomaly becomes more significant as various institutions create samples at different times, essentially ensuring a large variance in visual appearance.
- Annotation of training data is laborious and time consuming, and thus limited supervised data is available. Additionally, each new stain would require an equal investment to re-annotate and thus re-train.
- Precise reproducibility based on a wide range of input parameters is necessary for confidence and data exchange between operators. For an algorithm to become useful, institutions need to witness that the output created is less variant than intra-expert variability.
Contributions
- A tested hierarchical segmentation approach that marries an accelerated Mean Shift method and the Normalized Cuts method which we term Hierarchical Normalized Cuts (HNCut). A Matlab implementation of HNCut not only operates on large (1.5 million pixels) images in under 10 seconds, but is easily scalable to entire TMAs.
- Parameter insensitive segmentation for large images and the ability of HNCut to discriminate between regions with similar color values. The parameter for the Gaussian kernel in the affinity matrix of NCut is automatically computed. The parameters for the mean shift are automatically adjusted based on the variance of the output.
- Layman initialization of the system is possible, obviating the need for detailed ground truth annotation from an expert that is required for more sophisticated supervised classifiers.
Overview
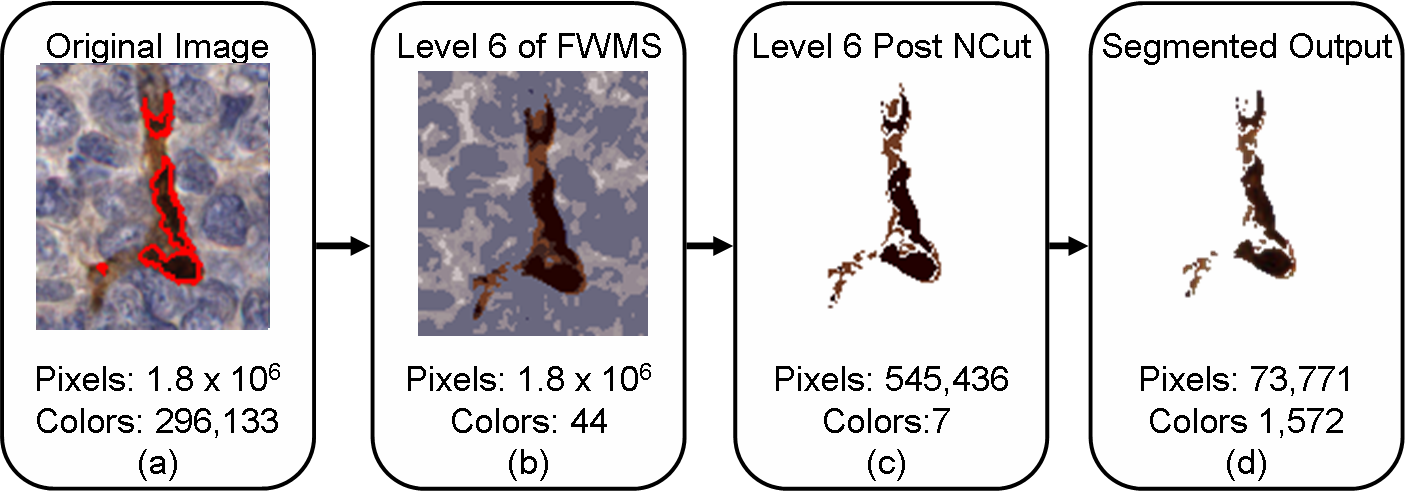
In the above flowchart, we display an image from our dataset (with ground truth marked in red) undergoing the HNCut procedure, with the intent of quantification of the vascular marker stain (brown color). The numbers shown in the boxes represent the reduced number of colors and pixels generated by the HNCut scheme at different levels of the heirarchy within a single cylinder (1500 x 1500 pixels, 300,000 colors) from a tissue micro array (TMA)
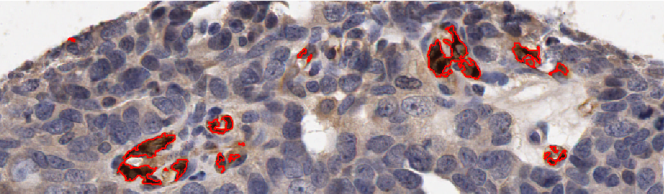
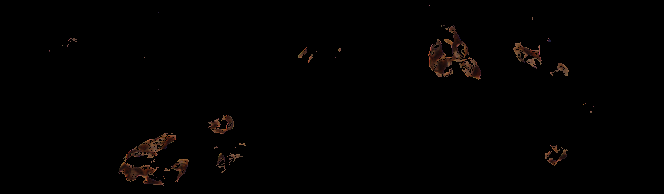
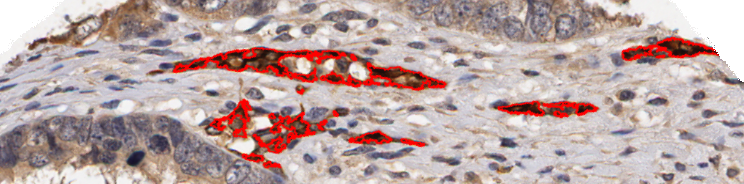

Two further examples of the HNCut algorithm working on Oca images. Again we can see the ground truth deliniated in red on the top image, with the respective HNCut output underneath.
Local Morphometric Scale
The major contribution of this piece of the thesis is to develop a system which can accurately classify pixels as being embedded in either tumor or stromal regions, and thus extendable to classifying lymphocytes as either TILs or non-TILs.Challanges
- Domain specific approaches require information about individual cells, such as size and dispersion pattern relative to its peers. The segmentation of individual cells is difficult due to clumping as a result of a three
dimensional tissue sample being scanned in two dimensions, and thus computationally expensive methods are needed for cell separation.
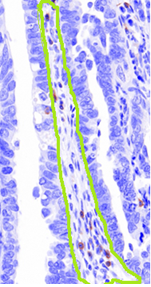
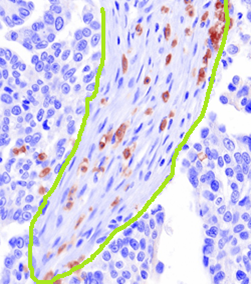
- Selection of an appropriate window size for standard approaches. We can see from the figure above, which has the stroma region manually circled in green, that although they are of the same class, the stark difference in size and shape between the two regions makes the selection of an appropriate window size or shape in a typical approach such as texture features is difficult.
- The stroma region is often nestled between areas of tumor, making not only its boundaries not clearly defined but the size of the associated region difficult to pre-determine.
Contributions
- A novel signature definition, which we term Local Morphometric Scale (LMS), allowing for quantitatively characterizing local heterogeneity. This is especially relevant in the context of histopathology which consists of notoriously heterogeneous images.
- The LMS yields a rotationally invariant, non-domain specific, quantitative signature at the pixel level which can be used for region classification, segmentation, and registration.
- This signature is accurate across a range of window sizes, overcoming common downfalls of texture and template matching based classifiers.
- The algorithm is well suited for GPU computing, allowing for a very high throughput.
- We develop a novel approach to the very important problem of separating out tumoral from stromal regions via application of LMS.
Overview
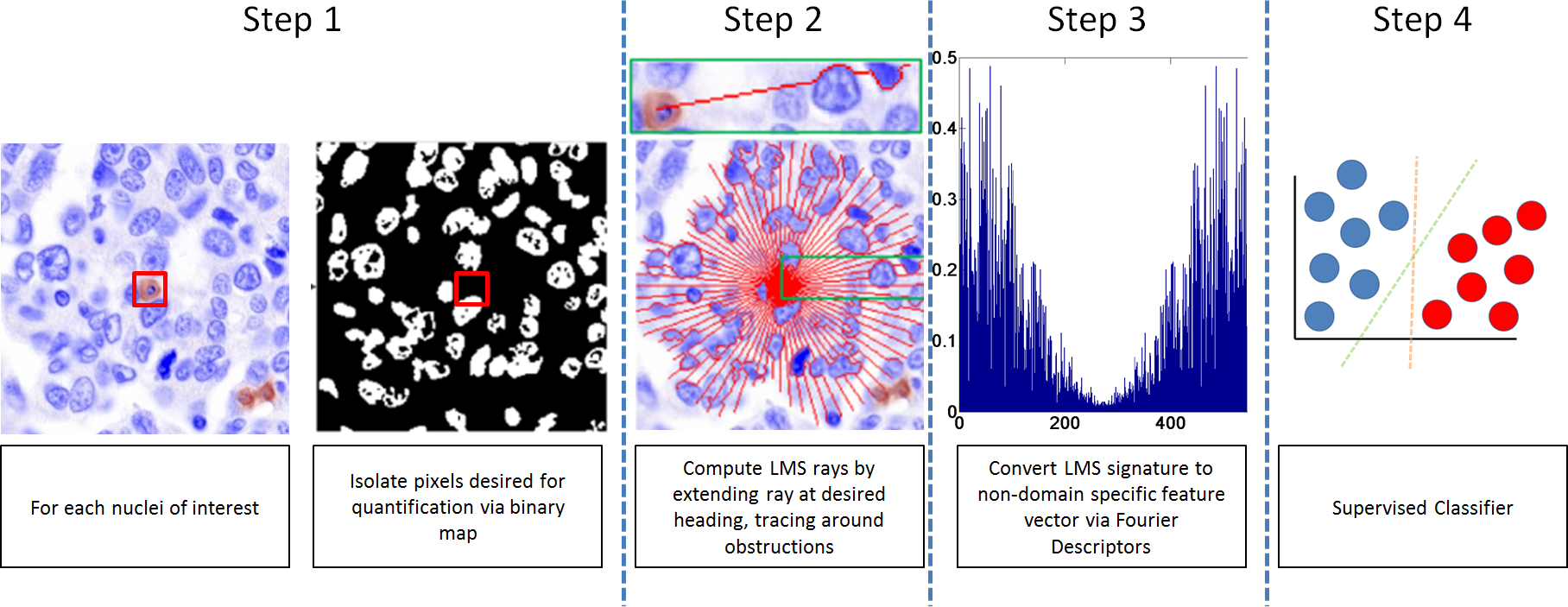
Overview of the LMS signature creation process. We can get the intuition that the more heterogeneous an area is the higher number of deviations from the straight line trajectory occur, on account of the rays attempting to take the path of least resistance and hence overcome obstacles along the way. On the other hand, the LMS signature will be smoother as a result of comprising fewer and smaller objects in homogeneous areas.
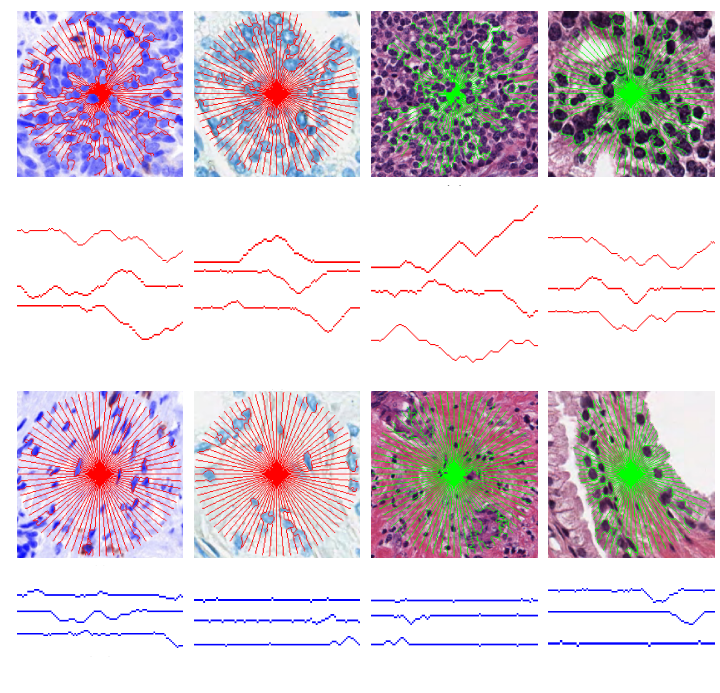
The LMS signature (in red or green) overlaid on a tumor regions (top) and benign regions (bottom) in an ovarian, prostate, breast, and prostate (different stain) images. Three rays are sampled from each image and presented beneath their respective images. We can see that in the non-tumor regions the LMS rays has fewer and smaller objects to obstruct its path, and thus the rays are less tortuous, unlike in the tumoral regions. Quantifying these differences allow for the classification of the regions using a supervised classifier.